Research Interests#
The multi-scale life of proteins#
Proteins function within extensive and highly interconnected networks, such that even slight changes in a protein caused by mutations can have far-reaching effects beyond its immediate connections. This propagation plays a key role in genetic diseases like cancer and neurodegenerative diseases, where a single mutation can affect the entire body. Better understanding of this propagation can improve the design of effective and minimally harmful treatments.
In many cases, individual proteins evolve to be multifunctional: their primary job is to organize the intersection of cellular networks. This ability to co-ordinate dissimilar cellular processes (such as metabolic or signaling pathways) is a promising feature for synthetic biologists when designing new living systems. However, our current understanding and tools for designing multifunctional proteins are limited. Therefore, my research focuses on developing these tools, using both experimental (wet lab) and computational (dry lab) techniques.
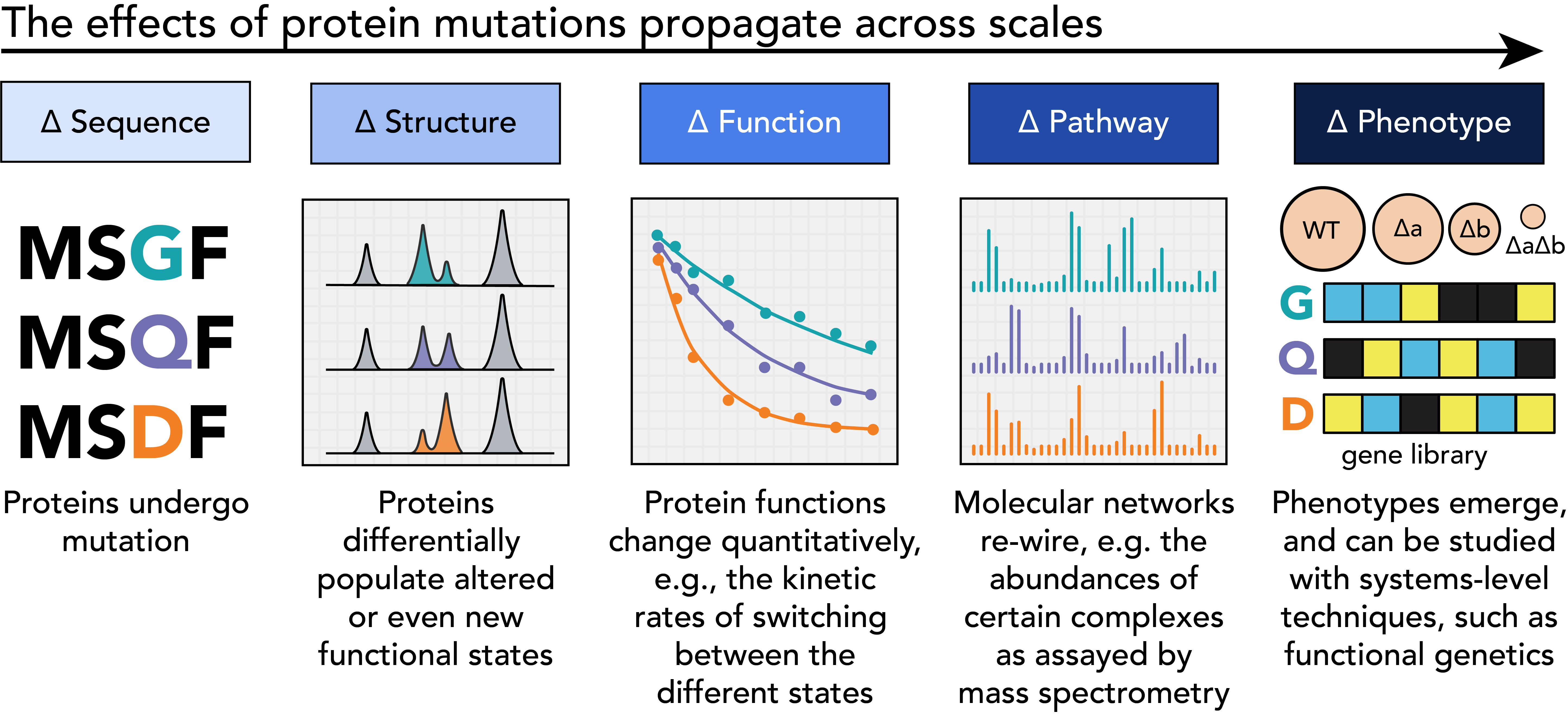
Postdoctoral work#
In my current position as a postdoctoral scholar in Lars Steinmetz’s lab, I am pursuing two research directions:
Engineering synthetic mitochondrial genomes in living cells. This involves solving longstanding issues in cloning toxic mtDNA species, as well as adapting nucleic acid delivery methods for integration of synthetic mtDNA in cultured human cells.
Incorporating cellular context into protein design workflows, by training deep learning models on data from combinatorial genomic screens.
Doctoral work#
In Tanja Kortemme’s lab, I studied how GTPases are allosterically regulated. GTPases are small proteins that act as “switches”, where they have an ON state and an OFF state determined by binding a GTP or GDP molecule. They control signaling by interacting with other proteins and activating cellular processes in a state-dependent manner. I studied:
How point mutations in one GTPase, Gsp1 (the yeast homolog of human Ran), can differentially and independently impact various processes in the cell.
How the cell might regulate these same processes by employing proteins that bind to Gsp1, making the same structural changes as the point mutations.
Whether we can discover the molecular mechanisms of allostery in Gsp1 using high-throughput mutational scans.
Undergraduate work#
I engineered growth factors for therapeutic applications in Jennifer Cochran’s lab. Growth factors binding to receptors on the cell surface tell the cell when to grow, for how long and how fast. This makes them key therapeutic targets to activate for wound healing, and to suppress in cancer. I studied how a cell’s response to growth factors depends on its specific combination of receptor proteins, and which internalization pathways are activated by each receptor. Specific mutations in the growth factors controllably induced distinct cellular responses, allowing for precise control over therapeutic efficacy.